2 Genotype and context data management with Poseidon
Poseidon is an open computational framework to enable standardised and FAIR (Wilkinson et al. (2016)) handling of genotypes with their highly relevant context information.
It includes a well-specified data format, advanced software tools, and public, community-maintained archives to support the entire archaeogenetic research cycle, from data acquisition to management, analysis and publication.
Poseidon and all its components is well documented at https://www.poseidon-adna.org. In this tutorial we will give a brief overview and highlight two workflows in the context of Poseidon.
2.1 The components of Poseidon
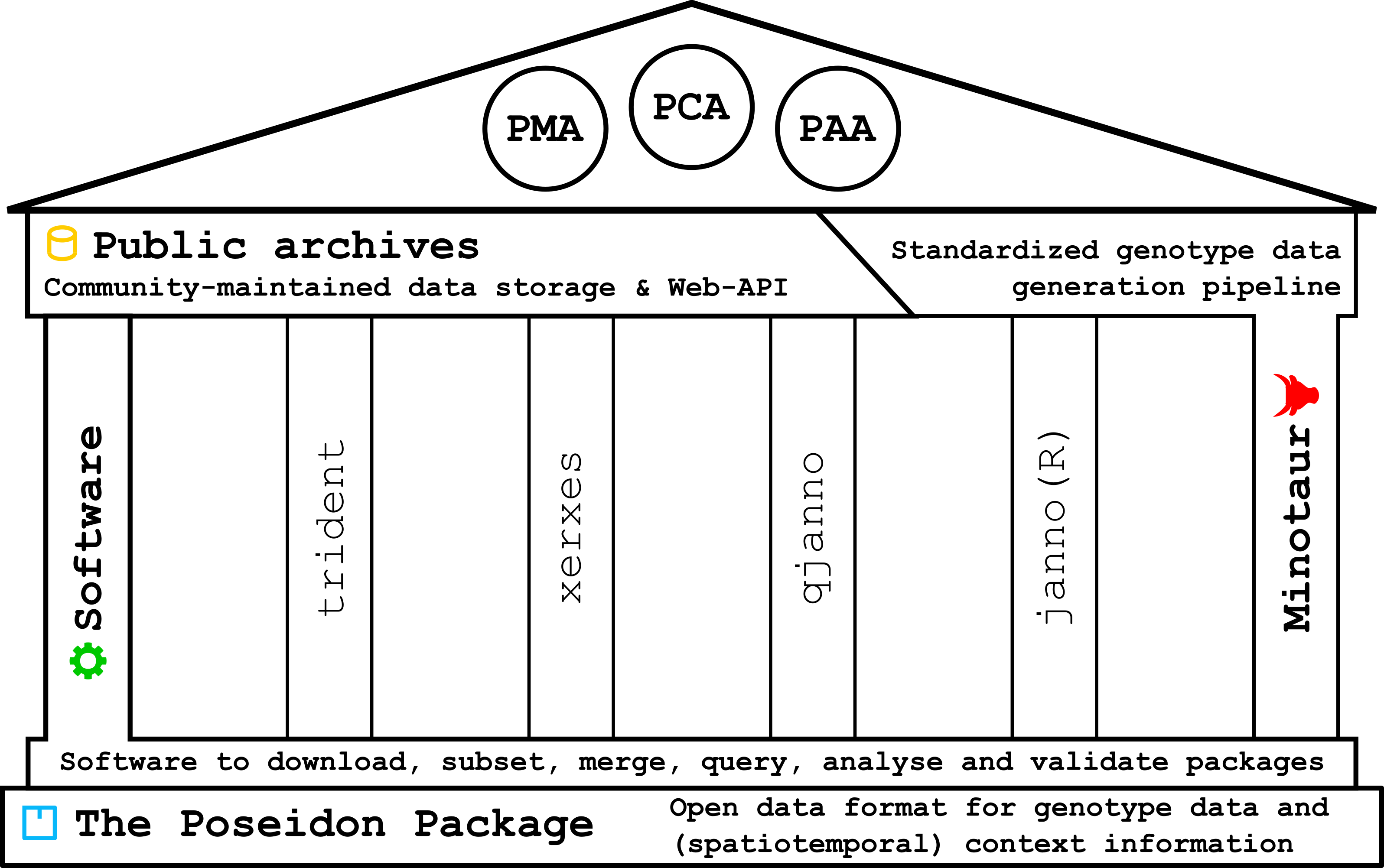
Poseidon is an entire ecosystem built on top of the data format specification of the Poseidon package.
2.1.1 The Poseidon package
A Poseidon package bundles genotype data in EIGENSTRAT/PLINK format with human- and machine-readable meta-data.
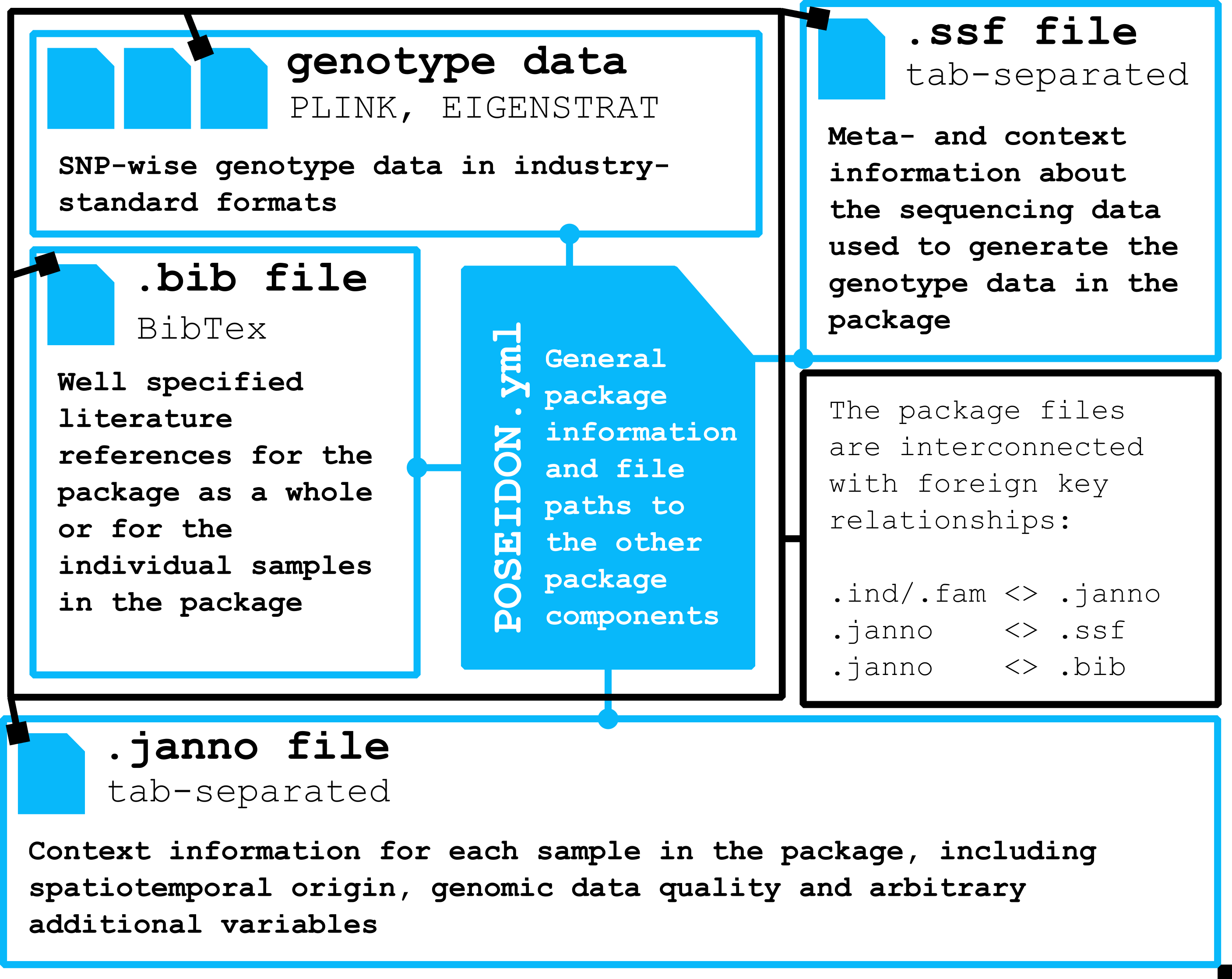
It includes sample-wise context information like spatio-temporal origin and genetic data quality in the .janno-file, literature in the .bib-file, and pointers to sequencing data in the .ssf-file.
.janno and .ssf have many predefined and specified columns, but can store arbitrary additional variables.
2.1.2 The software tools
trident is a command line application to create, download, inspect and merge Poseidon packages – and therefore the central tool of the Poseidon framework. The init subcommand creates new packages from genotype data, fetch downloads them from the public archives through the Web-API, and forge merges and subsets them as specified. list gives an overview over entities in a set of packages and validate confirms their structural integrity.
xerxes is derived from trident and allows to directly perform various basic and experimental genomic data analyses on Poseidon packages. It implements allele sharing statistics (\(F_2\), \(F_3\), \(F_4\), \(F_{ST}\)) with a flexible permutation interface.
janno is an R package to simplify reading .janno files into R and the popular tidyverse ecosystem (Wickham et al. (2019)). It provides an S3 class janno that inherits from tibble.
qjanno is another command line tool to perform SQL queries on .janno files. On start-up it creates an SQLite database in memory and reads .janno files into it. It then sends any user-provided SQL query to the database server and forwards its output.
2.1.3 The public archives
The Poseidon community maintains public archives for Poseidon packages to establish a central open point of access for published archaeogenetic genotype data.
- The Community Archive: Author supplied per-paper packages with the genotype data published in the respective papers. Partially pre-populated from various versions of the AADR.
- The AADR Archive: Complete and structurally unified releases of the Allen Ancient DNA Resource (Mallick et al. (2023)) repackaged in the Poseidon package format.
- The Minotaur Archive: Per-paper packages with genotype data reprocessed by the Minotaur workflow (see below).
The data is versioned with Git and hosted on GitHub for easy co-editing and automatic structural validation.
It can be accessed through a Web-API with various endpoints at server.poseidon-adna.org, e.g. /packages for a JSON list of packages in the community archive.
This API enables a little Archive explorer web app on the Poseidon website.
2.1.4 The Minotaur workflow
The Minotaur workflow is a semi-automatic workflow to reproducibly process published sequencing data from the International Nucleotide Sequence Database Collaboration (INSDC) archives into Poseidon packages.
Community members can request new packages by submitting a build recipe as a Pull Request against a dedicated submission GitHub repository. This recipe is derived from a Sequencing Source File (.ssf), describing the sequencing data for the package and where it can be downloaded.
Using the recipe, the sequencing data gets processed through nf-core/eager (Fellows Yates et al. (2021)) on computational infrastructure of MPI-EVA, using a standardised, yet flexible, set of parameters.
The generated genotypes, together with descriptive statistics of the sequencing data (Endogenous, Damage, Nr_SNPs, Contamination), are compiled into a Poseidon package and made available to users in the Minotaur archive.
2.2 Forging a dataset with trident
forge creates new Poseidon packages by extracting and merging packages, populations and individuals/samples from your Poseidon repositories. It can also work directly with your genotype data. In addition, forge allows merging of multiple data sets (packages), in contrast to mergeit which merges only two data sets at a time.
(-f/--forgeString) can be used to query entire packages, groups or individuals. In general --forgeString query consists of multiple entities, inside "" separated by , .
To include all individuals in a Poseidon package, use
*to surround the package title (e.g.*2019_Jeong_InnerEurasia*) . In cases where only genotype files are available, use the file name prefix.To include certain group(s) from a Poseidon package, simply add them to the
-fquery. No specific markers are required.Russia_HG_Karelia. You don’t have to specify the group, as trident will search all packages for the given group.To extract individuals only, surround them by
<and>.<ALA026>. To exclude individuals add package name*package*and<individual>with a dash sign."*2021_Saag_EastEuropean-3.2.0*,-<NIK003>"
trident forge \
-p pileupcaller.double.geno \
-d 2021_Saag_EastEuropean-3.2.0 \
-d 2016_FuNature-2.1.1 \
-f "*pileupcaller.double*,Russia_AfontovaGora3,<NIK003>" \
-o testpackage \
--outFormat EIGENSTRAT \
/forge has a an optional flag --intersect, that defines whether the genotype data from different packages should be merged with an intersect instead of the default union operation. The default is to output the union of all SNPs, by setting the additional SNPs from the other merged package as missing in the samples that did not have them originally. This option is useful for making a data set based on Human Origins (HO) SNPs for analysis like PCA and ADMIXTURE.
trident forge \
--intersect \
-p pileupcaller.double.geno \
-d 2012_PattersonGenetics-2.1.3 \
-o testpackage_HO \
--outFormat EIGENSTRAT \
/In case of PCA, --forgeFile can be used to merge necessary populations/groups from the available packages in the community archive to create specific PCA configurations.
trident forge \
-d /path/to/community/archive \
--forgeFile WestEurasia_poplist.txt \
-o WE_PCA \
/In addition, --selectSnps allows to provide forge with a SNP file in EIGENSTRAT (.snp) or PLINK (.bim) format to create a package with a specific selection. This option generates a package with exactly the SNPs listed in this file.
2.3 Contributing to the community archive

To maintain the public data archives, specifically the community archive and the minotaur archive, Poseidon depends on work donations by an interested community.
Many practitioners of archaeogenetics both produce genotype data from archaological contexts and require the reference data from other publications, provided in public archives, to contextualize it.
If authors themselves provide high-quality, easily accessible versions of their data beyond the raw data available at the INSDC databases, they gain at least three important advantages:
- Their work will be easily findable and potentially cited more often.
- They have primacy over how their data is communicated. That is, which genotypes, dates or group names they consider correct.
- Their results for derived, genotype based analyses (PCA, F-Statistics, etc.) can be reproduced exactly.
And the whole community wins, because sharing the tedious data preparation tasks empowers all researchers to achieve more in shorter time.

This tutorial explains the main cornerstones of a workflow to add a new Poseidon package to the community archive after publishing the corresponding dataset. The process is documented in more detail in a Submission guide on the website.
- Make yourself familiar with a number of core technologies. This is less daunting than it sounds, because: Superficial knowledge is sufficient and knowing them is useful beyond this particular task.
- Creating and validating Poseidon packages with the
tridenttool. - Free and open source distributed version control with Git.
- Collaborative working on Git projects with GitHub.
- Handling large files in Git using Git LFS.
- Create a package from your genotype data and fill it with a suitable set of meta and context information.
trident initallows to wrap genotype data in a dummy Poseidon package. Imagine we had genotype data for a number of individuals in EIGENSTRAT format:myData.ind
Sample1 M ExamplePop1 Sample2 F ExamplePop1 Sample3 M ExamplePop2myData.snp
rs3094315 1 0.020130 752566 G A rs12124819 1 0.020242 776546 A G rs28765502 1 0.022137 832918 T CmyData.geno
099 922 999With
trident init -p myData.geno -o myPackagewe can create a basic package around this data.$ ls myPackage myData.geno myData.snp myPackage.janno myData.ind myPackage.bib POSEIDON.ymlIn a next step we modify
POSEIDON.yml,.jannoand.bibto include the context information we consider relevant. All of these files are well specified and documented, so we only demonstrate a minimal change for this example:We replace the main contributor for the package.
myPackage/POSEIDON.yml
poseidonVersion: 2.7.1 title: myPackage description: Empty package template. Please add a description contributor: - name: Clemens Schmid #- name: Josiah Carberry email: clemens_schmid@eva.mpg.de # email: carberry@brown.edu orcid: 0000-0003-3448-5715 # orcid: 0000-0002-1825-0097 packageVersion: 0.1.0 lastModified: 2023-10-18 genotypeData: format: EIGENSTRAT genoFile: myData.geno snpFile: myData.snp indFile: myData.ind snpSet: Other jannoFile: myPackage.janno bibFile: myPackage.bibWhen we applied all necessary modifications we can confirm that the package is still valid with
trident validate -d myPackage.
- Submit the package to the community archive.
- To submit the package we have to create a fork of the community archive repository on GitHub. This requires a GitHub account.
And then clone the fork to our computer, while omitting the large genotype data files. Note that this requires several setup steps to work correctly:
- Git has be installed for your computer (see here)
- You must have created an ssh key pair to connect to GitHub via ssh (see here)
- Git LFS has to be installed (see here) and configured for your user with
git lfs install
GIT_LFS_SKIP_SMUDGE=1 git clone git@github.com:<yourGitHubUserName>/community-archive.gitWith the cloned repository on our system we can copy the files into the repositories directory and commit the changes.
in the community-archive directory
cp -r ../myPackage myPackage git add myPackage git commit -m "added a first draft of myPackage" git pushIn a last step we can open a Pull Request on GitHub from our fork to the original archive repository. Poseidon core members will take it from here.